Blog More than a hundred elite rice lines for LAC, from the rice breeding programs (Alliance-FLAR) to be adopted by all

By Nicolas Hoyos, Robert Andrade
Year by year hundreds of elite rice lines are developed by the Alliance of Bioversity International and CIAT and FLAR breeding programs, but “how much diversity do we have?” and “what is the most efficient way study it?” are remaining questions of relevance.
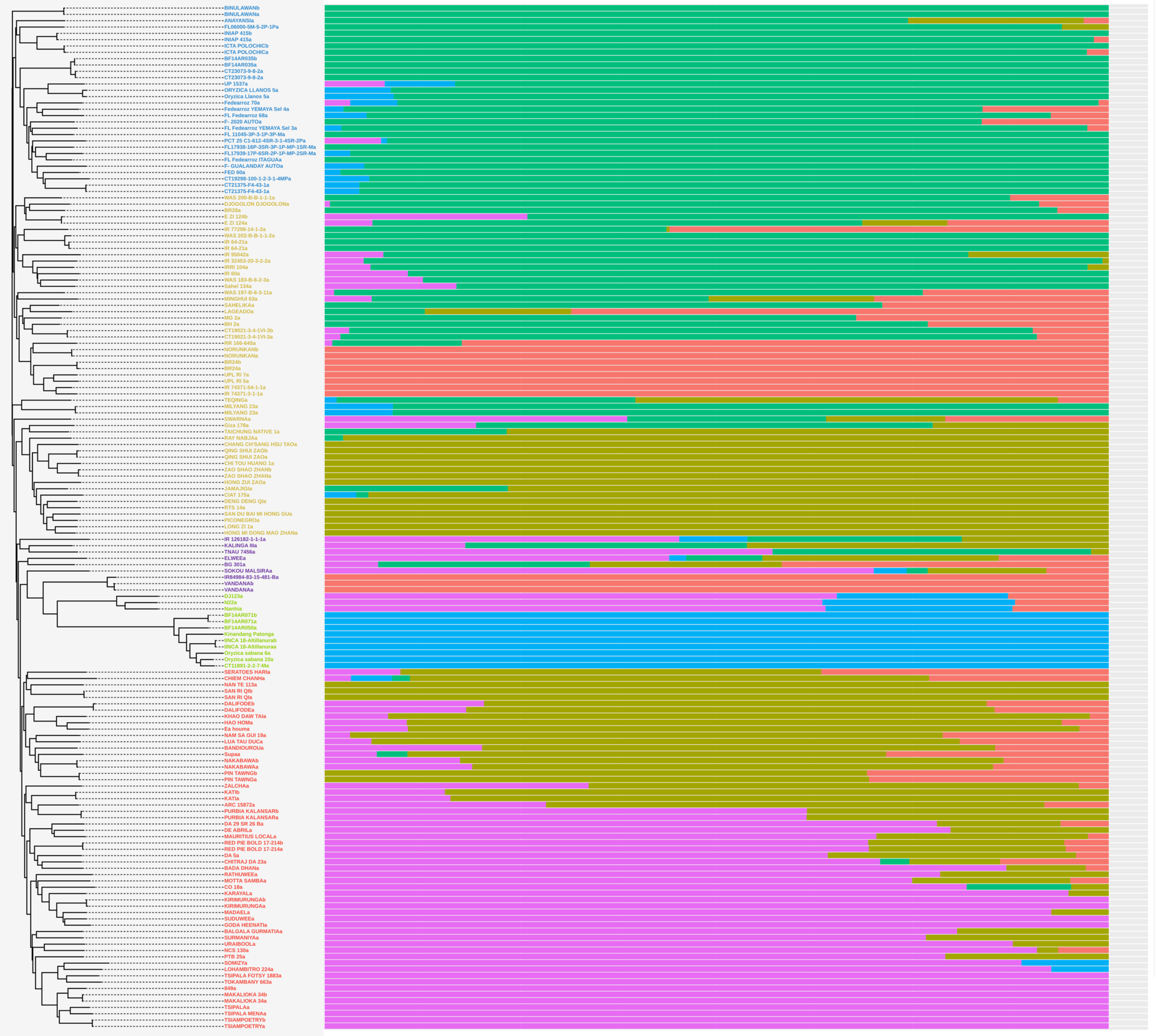
Genomics offers a huge variety of possibilities for both research and crop improvement purposes; prediction of relevant agricultural traits, genotype-phenotype interactions, and directed breeding just to name a few. For this reason, the rice breeding programs decided to perform full sequencing on 137 elite rice cultivars and perform diversity and population genetics analysis. We determined that due to the extensive history of crosses within different lines, a tree structure is inadequate to show the relationships between our lines, and admixture analysis provides a more accurate picture of their relatedness.
The used methodology consisted on sequencing with a mean depth from 25x to more than 60x, but our results showed that most variants were already determined at 14x; our first recommendation is that unless deep structural analysis or repeats copy number is to be studied, the target sequencing depth should be of no more that 20x, as resources would be more efficiently used in analyzing more lines rather than achieving higher resolution via deeper sequencing.
To take advantage of a wider context provided by the vast knowledge available for rice genomics we enriched our dataset with lines selected from the 3,000 genomes project (The 3,000 rice genomes project 2014), as it provides comprehensive information on the whole diversity of cultivated rice around the world. When making neighbor joining trees, we realized that the resulting branches were very unstable and extremely short in length, most likely because of very recent crosses. Also observed by many hybrid individuals when grouping using admixture, which showed much more stability and was consistent with and without enrichment with 3,000 genomes data.
Some of our most interesting results lie within the diversity analysis with pi diversity statistic and MAF curves. We compared observed diversity between our panel and that found in the 3,000 genomes project, which is representative of global diversity. We found that for Indica, even if the diversity is lower, the lines we analyzed had a pi statistic very similar to the global diversity; while for Japonica and Aus gene pools it was remarkably lower. Implying that bringing in lines from these origins could greatly increase available diversity.
While our study provides insightful results, there is still much work to be done, we seek to perform introgression analysis to pinpoint sections of the genome that have transferred between groups. To use phenotypic characterization and perform GWAS to look for variants related to agriculturally relevant traits. To use adoption data to calculate the production increase generated by the rice improvement program, taking in account that previous studies estimated that more than two thirds of all rice area sowed is sowed with some material related to the rice breeding programs of CIAT and/or FLAR (Yamano et al., 2016) (Labarta et al., 2017). And to keep adding variants to our database, to get a better idea of not only what we have achieved, but also the future potential and most effective ways to exploit it. Cooperation between breeders, geneticist, farmers, seed producers, industry and agricultural development economist is paramount to make best use of the genetic resources we must strengthen rice production in a region that by decades has maintain growth rates in their average yields (Mishra, et al. 2022).
References:
- The 3,000 rice genomes project. (2014). GigaScience, 3(1). https://doi.org/10.1186/2047-217x-3-7
- Labarta, R., Andrade, R., Salazar, D. M., Rivera, T., Orrego, M., & Pinillos, J. (2017). The Impacts of CIAT’s Collaborative Research. International Center for tropical Agriculture (CIAT). Cali, CO. 12 p.
- Mishra, A. K., Pede, V. O., Arouna, A., Labarta, R., Andrade, R., Veettil, P. C., Bhandari, H., Laborte, A. G., Balie, J., & Bouman, B. (2022). Helping feed the world with Rice Innovations: CGIAR research adoption and socioeconomic impact on farmers. Global Food Security, 33, 100628. https://doi.org/10.1016/j.gfs.2022.100628
- Yamano, T., Arouna, A., Labarta, R. A., Huelgas, Z. M., & Mohanty, S. (2016). Adoption and impacts of International Rice Research Technologies. Global Food Security, 8, 1–8. https://doi.org/10.1016/j.gfs.2016.01.002
