MusaDeepMosaic: Development of a machine learning genomic mosaic classifier tool.
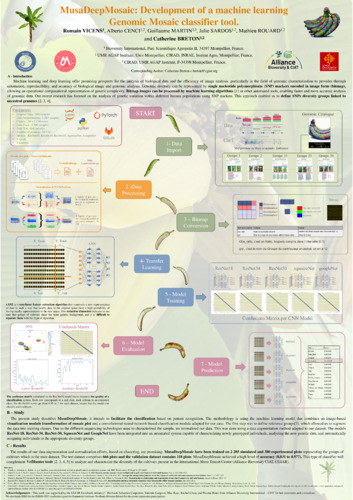
Machine learning and deep learning offer promising prospects for the analysis of biological data and the efficiency of image analysis, particularly in the field of genomic characterization to provides through automation, reproducibility, and accuracy of biological image and genomic analysis. Genomic diversity can be represented by SNP markers encoded in image form (bitmap), allowing an operational computational representation of genetic complexity. Bitmap images can be processed by machine learning algorithms [1] or other automated tools, enabling faster and more accurate analysis of genomic data.
Our recent research has focused on the analysis of genetic variation within different banana populations using single nucleotide polymorphisms (SNPs) markers. This approach enabled us to define SNPs diversity groups linked to ancestral genomes [2, 3, 4]. The genome of cultivated varieties were then visualized as colored mosaics, which provided a unique visual representation of the genetic complexity of the banana genome ancestry. All cultivated varieties display a composite chromosomic structure with a complex mosaic of segments from different wild species and sub-species that was curated into groups [6]. However, the precise definition of these groups and their attribution to specific ancestors requires expert manual work.
The present study describes MusaDeepMosaic intends to facilitate the classification based on pattern recognition. The methodology is using the machine learning model that combine an image-based visualization module transformation of mosaic plot and a convolutional neural network-based classification module adapted for our case. The first step was to define reference groups, which allowed us to segment the data into training classes. However, to improve the accuracy of our model, we needed to significantly increase our data set. Due to the different sequencing technology, we normalized our data. This was done using a data augmentation method adapted to our dataset, which allowed us to expand our data corpus without compromising quality. The ResNet-50 model, a 50-layer deep convolutional neural network introduced in 2015 for image recognition, was utilized in this study. Optimized for accurate performance and fast processing times, ResNet-50 will be integrated into an automated system capable of characterizing newly genotyped individuals, analyzing the new genetic data, and automatically assigning individuals to the appropriate diversity groups.
The initial results of our data augmentation and normalization efforts, based on clustering, are encouraging. MusaDeepMosaic will be trained on 1,483 simulated and 317 experimental plots representing the groups of cultivars which is the train dataset. The test dataset comprised 200 plots and the validation dataset contain 178 plots. MusaDeepMosaic achieved a higher level of accuracy (0,97 to 1). This type of classifier will complement VcfHunter tools [2, 3, 4, 5] to analyze and characterize the diversity of the cultivars present in the International Musa Transit Center (Alliance Bioversity CIAT, CGIAR).