Let's show Structural Variations
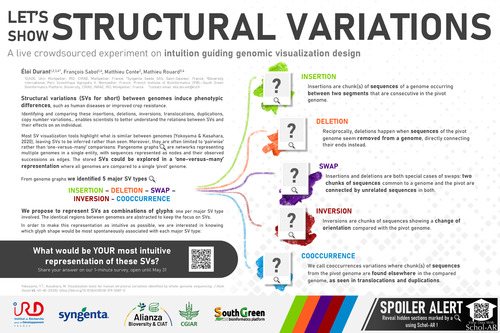
Genomes can be subject to Structural Variations (SVs): rearrangements of more than 50kb of a sequence of nucleotides. These variations can have various effects depending on where they take place in a genome (for example within a gene or a control region) and influence the phenotypes. Comparing these SVs between genomes enables scientists to better understand the links between the rearrangements and their effects. However there are few tools that offer a visual exploration of these SVs, as most of them focus only on the sequences that are common between two genomes. We propose a visual representation of these SVs as a glyphs integrated within a matrix-like display, based on the divergences between to path / genomes within pangenome graphs. One genome acts as a reference coordinate system (the ‘pivot genome’), and all other genomes are then compared directly to it using the glyphs to highlight the differences. In order to create the most intuitive (therefore easiest to understand) glyphs for each type of SVs, we encourage the VIZBI community to answer a 1-minute survey and discover our proposed glyphs.